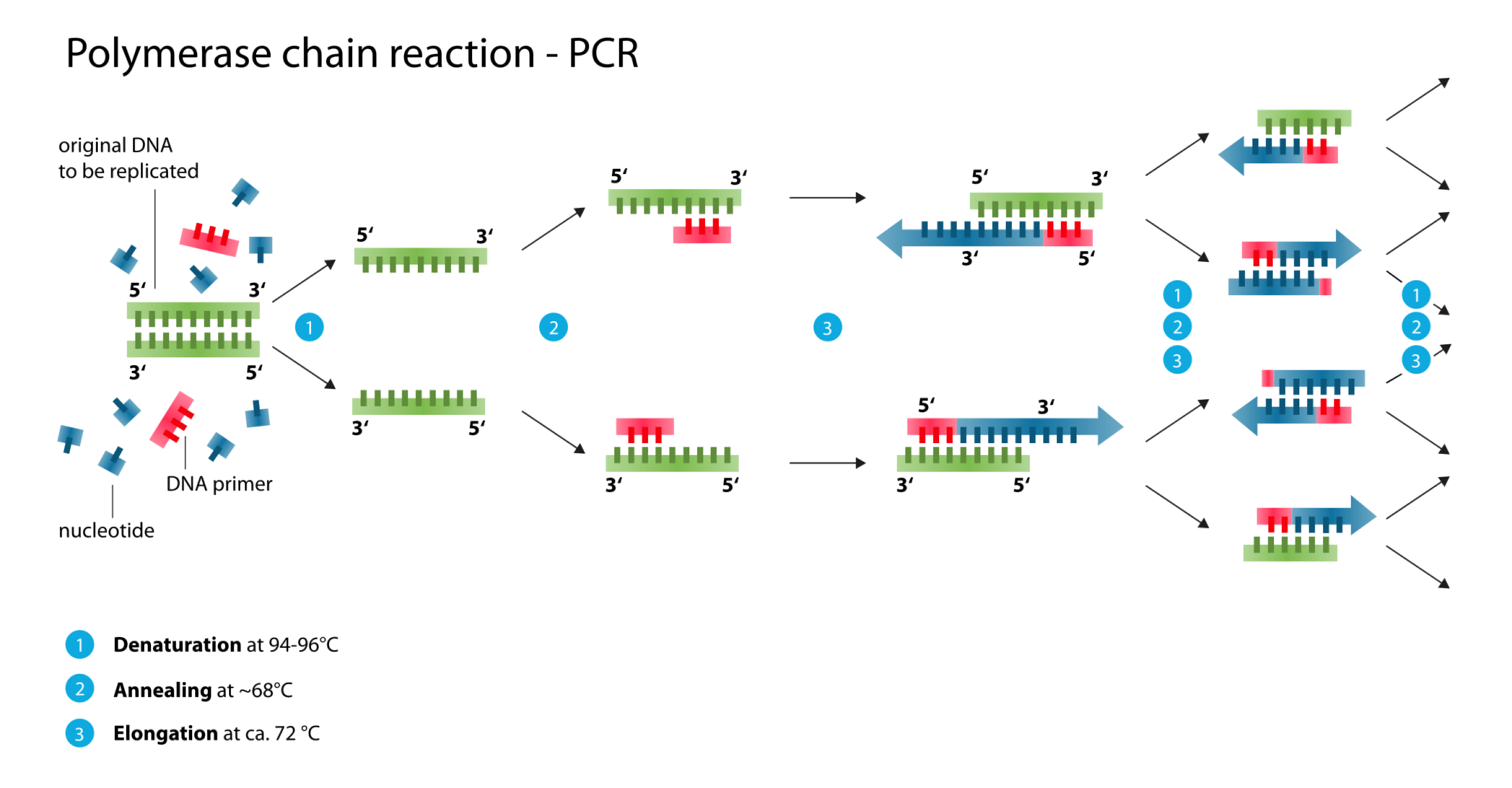
Polymerase chain reaction (PCR) is an efficient method for greatly amplifying the quantity of DNA templates. Taq polymerase is frequently used in the polymerase chain reaction (PCR), which is a thermostable DNA polymerase named after the thermophilic bacterium Thermus aquaticus from which it was originally isolated in 1976.
STEP 1: Prepare DNA template
Usually, for plasmid DNA, 1-10 ng; for genomic DNA, 50-500 ng per reaction is needed. Normally, DNA template does not need to be purified. However, both purity and the amount of template can strongly influence the outcome of the reaction.
STEP 2: Design primer
Generally, primers used are 18-23 mer in length. Use Primer3 free online software or other assistant tools like Primer Premier to design proper primers. Choose the best primer pairs from those options according to the parameters, such as size, TM, GC%, etc.
STEP 3: Determine annealing temperature:
Melting temperature (Tm) of primers can be calculated by the following formula:
Tm = [[(TmForward +TmReverse)×1/2]-10]°C.
Tm°C is a good annealing temperature to start with. However, optimal annealing temperatures can only be determined experimentally for a certain primer/template combination. Temperature gradient PCR is often a way to finalize an optimal annealing temperature.
STEP 4: For a 50 μl reaction, add:
|
10x PCR buffer |
5 μl |
|
DNA template |
X μl |
|
Forward Primer |
1 μl |
|
Reverse Primer |
1 μl |
|
dNTPs Mix |
1 μl |
|
Taq Polymerase |
0.25μl |
|
Sterile ddH2O |
To 50μl |
Notes:
1.Primers diluted to working concentration (10µM working stocks are sufficient for most assays).
2.Final Concentration of primers should be 0.1-0.5 µM, final Concentration of dNTPs Mix should be 200µM, final concentration of DNA template should be 200 pg/µL
3. Mix gently by vortex and briefly centrifuge to collect all components to the bottom of the tube.
STEP 5: A typical PCR program
|
CYCLE STEP |
TIME |
TEMP |
CYCLES |
|
Initial denaturation |
30 sec |
95 °C |
1 |
Denaturation
Annealing
Extension |
15-30 sec
15 sec
depends on length, 1 min/ kb |
95 °C
depends on Tm
72℃ |
30 |
|
Final extension |
5 min |
72 °C |
1 |
|
Keep sample |
∞ |
4 °C |
- |
Notes:
The amplification parameters will vary depending on the primers and the thermal cycler used. It may be necessary to optimize the system for individual primers, template, and thermal cycler.