Construction and Packaging Service of virus Vector
Construction and Packaging Services of AAV
Large scale AAV production
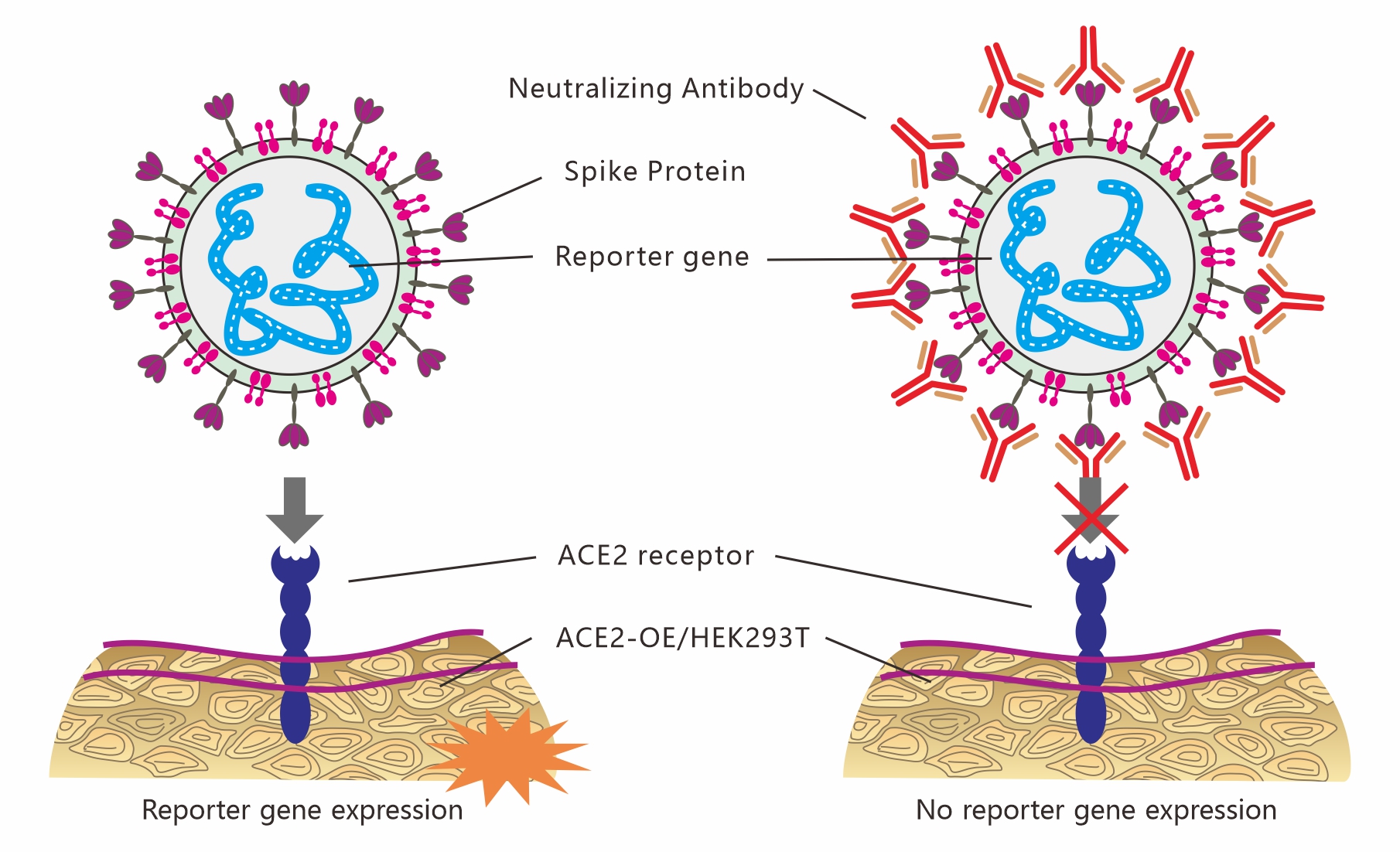
COVID-19 (SARS-CoV-2) Pseudovirus
|
Cat. No.
|
Product Name
|
Description
|
|
SARS-CoV-2-V03001
|
PV-SARS-CoV-2-S-VSV-
△G-EGFP |
VSV Backbone, SARS-CoV-2 Spike Protein (WT)-EGFP
|
|
SARS-CoV-2-V03005
|
PV-SARS-CoV-2-S-VSV-
△G-Luciferase |
VSV Backbone, SARS-CoV-2 Spike Protein (WT)-Luciferase
|
|
SARS-CoV-2-LV-0046
|
PV-SARS-CoV-2-S-LV-CMV-mcherry
|
HIV-1 Backbone,SARS-CoV-2 Spike Protein (WT)-mcherry
|
|
SARS-CoV-2-LV-0052
|
PV-SARS-CoV-2-S-LV-CMV-EGFP
|
HIV-1 Backbone, SARS-CoV-2 Spike Protein (WT)-EGFP
|
|
SARS-CoV-2-LV-0588
|
PV-SARS-CoV-2-S-LV-CMV-Luciferase
|
HIV-1 Backbone, SARS-CoV-2 Spike Protein (WT)-Luciferase
|
|
SARS-CoV-2-V03002
|
PV-SARS-CoV-2-S-(D614G)-VSV-△G-mcherry
|
VSV Backbone, SARS-CoV-2 Spike (D614G)-mcherry
|
|
SARS-CoV-2-K-LV-0588
|
PV-SARS-CoV-2-S-(D614G)-rLV-CMV-Luciferase
|
HIV-1 Backbone, SARS-CoV-2 Spike (D614G)-Luciferase
|
|
SARS-CoV-2-A-V03002
|
PV-SARS-CoV-2-S (B1.1.7) -VSV-△G-mCherry
|
VSV Backbone, SARS-CoV-2 Spike Protein (UK B.1.1.7;:
N501YA570D,P681H,T716I,S982A,D1118H)-mCherry |
|
SARS-CoV-2-K-LV-0588
|
PV-SARS-CoV-2-S(B.1.1.7)-rLV-CMV-Luciferase
|
HIV-1 Backbone, SARS-CoV-2 Spike Protein (UK B.1.1.7;:
N501YA570D,P681H,T716I,S982A,D1118H)-Luciferase |
|
SARS-CoV-2-B-V03002
|
PV-SARS-CoV-2-S(SA-B.1.351) -VSV-△G-mCherry
|
VSV Backbone, SARS-CoV-2 Spike Protein
(South African variant Δ3 B.1.351, 20H/501Y.V2, Beta; L18F, D80A, D215G, ΔL242/A243/L244, R246I, K417N, N501Y, E484K, D614G, A701V)-mCherry |
|
SARS-CoV-2-B-LV-0588
|
PV-SARS-CoV-2-S(SA-B.1.351)-LV-CMV-Luciferase
|
HIV Backbone, SARS-CoV-2 Spike Protein(
South African variant Δ3 B.1.351, 20H/501Y.V2, Beta; L18F, D80A, D215G, ΔL242/A243/L244, R246I, K417N, N501Y, E484K, D614G, A701V)-Luciferase |
|
V04006
|
PV-SARS-CoV-2-S(501Y.V2)-VSV-△G-mCherry
|
VSV Backbone, SARS-CoV-2 Spike (
South African variant B.1.351, 20C/501Y.V2; L18F, D80A, D215G, L242H, R246I, K417N, N501Y, E484K, D614G, A701V)-mCherry |
|
SARS-CoV-2-K-LV-0588
|
PV-SARS-CoV-2-S(501Y.V2)-rLV-CMV-Luciferase
|
HIV Backbone, SARS-CoV-2 Spike (
South African variant B.1.351, 20C/501Y.V2; L18F, D80A, D215G, L242H, R246I, K417N, N501Y, E484K, D614G, A701V)-Luciferase |
|
V04007
|
PV-SARS-CoV-2-S(B1.1.617)-VSV-△G-mCherry
|
VSV Backbone, SARS-CoV-2 Spike (
Indian variant B.1.617, RBD mutations only; L452R, E484Q, D614G)-mCherry |
|
SARS-CoV-2-K-LV-0588
|
PV-SARS-CoV-2-S(B1.1.617)-rLV-CMV-Luciferase
|
HIV Backbone, SARS-CoV-2 Spike (
Indian variant B.1.617, RBD mutations only; L452R, E484Q, D614G)-Luciferase |
|
SARS-CoV-2-D-LV-0588
|
PV-SARS-CoV-2-S(IN-B.1.617.2)-LV-CMV-Luciferase
|
HIV Backbone, SARS-CoV-2 Spike (
Indian variant B.1.617.2; T19R, G142D, del156/157, R158G, L452R, T478K, D614G, P681R, D950N)-Luciferase |
|
SARS-CoV-2-K-LV-0588
|
PV-SARS-CoV-2-S(IN-B.1.617.1)-LV-CMV-Luciferase
|
HIV Backbone, SARS-CoV-2 Spike (Indian variant B.1.617.1;
G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H)-Luciferase |
|
SARS-CoV-2-G-LV-0588
|
PV-SARS-CoV-2-S(BZ-P.1)-LV-CMV-Luciferase
|
HIV Backbone, SARS-CoV-2 Spike (Brazilian variant:
L18F,T20N,P26S,D138Y,R190S,K417T,E484K, N501Y,D614G,H655Y,T1027I)-Luciferase |
|
SARS-CoV-2-G-LV-0588
|
PV-SARS-CoV-2-S(B.1.1.529)-LV-CMV-Luciferase
|
HIVBackbone, SARS-CoV-2 Spike(A67V, Δ69-70, T95I,
G142D, Δ143-145, Δ211 212, ins214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F) |
Recombinant viral express SARS-COV-2-S, ACE2 and TMPRSS2 protein
| Vector | Cat. | Vector Name | Biosafety Level |
| Lentiviral vector | LV-0537 | rLV-CMV-SARS-CoV-2-S-2A-mCherry-WPRE | BSL-2 |
| LV-0538 | rLV-Ef1a-ACE2-2A-EGFP-WPRE | ||
| LV-0539 | rLV-Ef1a-ACE2-2A-EGFP--CMV-Puro-WPRE | ||
| LV-0558 | rLV-CMV-TMPRSS2-2A-mCherry-WPRE | ||
| LV-0559 | rLV-CMV-ACE2-2A-TMPRSS2-2A-EGFP-WPRE | ||
| AAV vector | PT-2701 | rAAV-CMV-TMPRSS2-2A-BFP-WPRE | |
| PT-2702 | rAAV-CMV-SARS-CoV-2-S-2A-mCherry-WPRE | ||
| PT-2703 | rAAV-Ef1a-ACE2-2A-EGFP-WPRE |